Introduction
Clinical studies of patients are stored in electronic health records (EHR) such as clinical letters or discharge summaries, consisting primarily of unstructured text. Named Entity Recognition (NER), is a fundamental task in Natural Language Processing(NLP) to classify mentioned words into predefined entities such as name, person, organization, etc. In the biomedical domain, this technique takes care of entities of interest such as diseases, drugs, symptoms, etc. Legacy systems built for information extraction rely primarily on structured data, and are unable to extract such hidden information from unstructured medical texts. Considering the complexity of medical text, a deep learning based supervised learning is used to tackle such entities. We use Scispacy, a Python package for processing biomedical, scientific or clinical text[1].
Scispacy
Scispacy is the scientific version of spacy, a text processing tool used for various NLP tasks such as tokenization, entity tagging, part of speech tagging, etc. Scispacy contains models trained on biomedical domain, that can be used for named entity recognition, dependency parsing, sentence segmentation, etc[2].
Approach: The overall approach for NER is schematically shown in Figure 1.
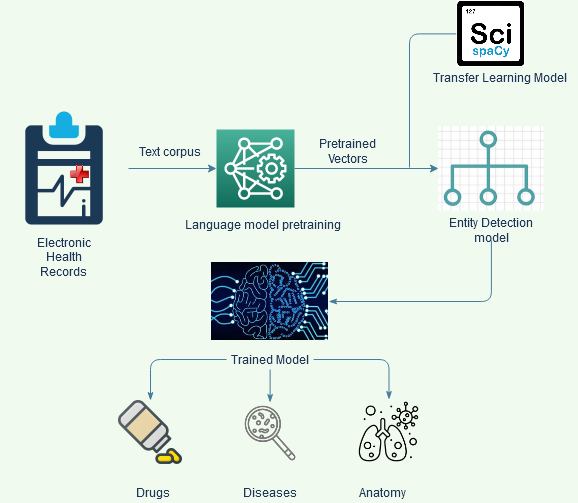
Fig. 1 Schematic representation of the NER approach
The data for entity extraction is sourced from freely available datasets, containing labelled entities. The diseases, anatomy and drugs are trained separately using NCBI, AnatEM and BC5CDR datasets respectively.
Once we have the labelled data, we use the spacy NER module to train our custom model[3].
Spacy NER uses a novel bloom embedding strategy with sub word features, used to support huge vocabularies in tiny tables. Convolutional Neural Network(CNN) layers are used with attention mechanisms, to give better efficiency than the widely used LSTM or BiLSTM architectures. The models tend to work well on smaller datasets and are much faster than a sequential Recurrent Neural Network (RNN) or LSTM model.
The deep learning technique ensures that not only the meaning of the word is used to predict the entities, but also the context of the surrounding words in the sentence is being taken into account for predicting our entities of interest.
Considering the complexity and diversity of biomedical entities, any model requires a large amount of data to successfully build the required connections to accurately predict each entity. There are tens of thousands of drugs that may not be included in our training data. Training a model on a limited vocabulary will restrict the scope of the model.
For example:
“The patient has an allergy to penicillin.”
In the above line, if the model does not contain the vocabulary of penicillin, it may not be correctly identified as a drug. To overcome such problems, we use transfer learning with the help of Scispacy. Scispacy contains trained models on biomedical domain, with an expanded vocabulary and their trained embeddings. We use them as a base model, to significantly improve our predictions and make it more robust.
For further improvement, we train a language model to learn word vectors, before the actual model training begins. To pretrain, we use the entire combined text corpus that is kept aside for training and validation for all 3 entities. This additional step trains word vectors on our training texts, to gather the context according to their surrounding words. Instead of initializing the neural network layer with random weights, and learning the weights by training, we begin this step having the learned vectors from the language model. The combination of the extended vocabulary and embeddings from Scispacy and the pretrained vectors provides a better result post training.
Figure 2 visualizes the model prediction on an unlabelled text snippet from an electronic health record (EHR).

Summary
The model outperforms unsupervised approaches such as dictionary matching or even sequential deep learning models such as BiLSTM, with limited training data availability. The results are proportional to the approach described in the paper[4] where retrained Scispacy with pretrained vectors provided the best F1 score. Scientific research in the medical domain has a rich literature comprising millions of health records, clinical letters, research papers, etc. However, due to their unstructured nature, applications such as an entity extractor can help researchers and medical professionals to extract valuable information and further process them to some structured format. Adverse or Positive entity relationships such as drugs-diseases or protein-DNA can be studied once extracted[5]. Other downstream tasks such as negation detection, tense detection and knowledge discovery are also possible. The approach used is not limited to the healthcare domain, and can be extended to insurance, retail, legal, and other domains. For example, one potential application can be to extract legal entities such as captions, past judgements, specific laws, etc from legal corpus.
We, at AlgoAnalytics, have built the application for MediTerm Extraction using custom Scispacy. For a detailed demo please visit : https://textsense.onestop.ai/demo/dashboard/meditermExtraction
Demo and Contact Info:
To check our other demos, please visit: https://onestop.ai
For further information, please contact: info@algoanalytics.com
References:
[1]https://allenai.github.io/scispacy/
[2]https://www.aclweb.org/anthology/W19-5034.pdf#cite.Lample2016NeuralAF
[4]https://arxiv.org/ftp/arxiv/papers/1910/1910.11241.pdf
This post first appeared in Medium

